Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 17
Few-Shot Classification with Multiscale Feature Fusion for Clinical Skin Disease Diagnosis
Received 1 February 2024
Accepted for publication 22 April 2024
Published 6 May 2024 Volume 2024:17 Pages 1007—1026
DOI https://doi.org/10.2147/CCID.S458255
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Rungsima Wanitphakdeedecha
Tianle Chen, Qi Liu, Jie Yang
College of Biomedical Engineering, Sichuan University, Chengdu, 610065, People’s Republic of China
Correspondence: Qi Liu, College of Biomedical Engineering, Sichuan University, Chengdu, 610065, People’s Republic of China, Email [email protected]
Introduction: Skin disease is one of the most common diseases and can affect people of all ages and races. However, the diagnosis of skin diseases via observation is a highly challenging task for both doctors and patients, and would benefit from the use of an intelligent system. Building a large benchmark with professional dermatologists is resource-intensive, and we believe that few-shot learning (FSL) methods would be helpful in solving the problem of annotated data scarcity. In this paper, we propose CDD-Net (Context Feature Fusion and Dual Attention Dermatology Net), a plug-in module for FSL clinical skin disease classification.
Methods: Current FSL methods used in skin disease classification are limited to nonuniversal approaches and few disease classes. Our CDD-Net has a flexible structure, including a context feature–fusion module and dual-attention module to extract discriminating texture feature and emphasize contributive regions and channels. The context feature–fusion module localizes discriminatory texture details of skin lesions by integrating features from different layers, while the dual-attention module highlights discriminative regions via channel-wise and pixel-wise depictions based on weight vectors and restrains the contributions of irrelevant areas. We also present Derm104, a new clinical skin disease data benchmark that has significant coverage of rare diseases and reliable annotation between primary species and subspecies for better validation of our approach.
Results: Our experiments validated the versatility of CDD-Net for different FSL methods and achieved an improvement in accuracy of up to 9.14 percentage points compared with the vanilla network, which can be considered state of the art. The ablation study also showed that the dual-attention module and context feature–fusion module worked efficiently in CDD-Net.
Keywords: clinical skin image, computer-aided diagnosis, few-shot learning, feature fusion, attention mechanism
Graphical Abstract:
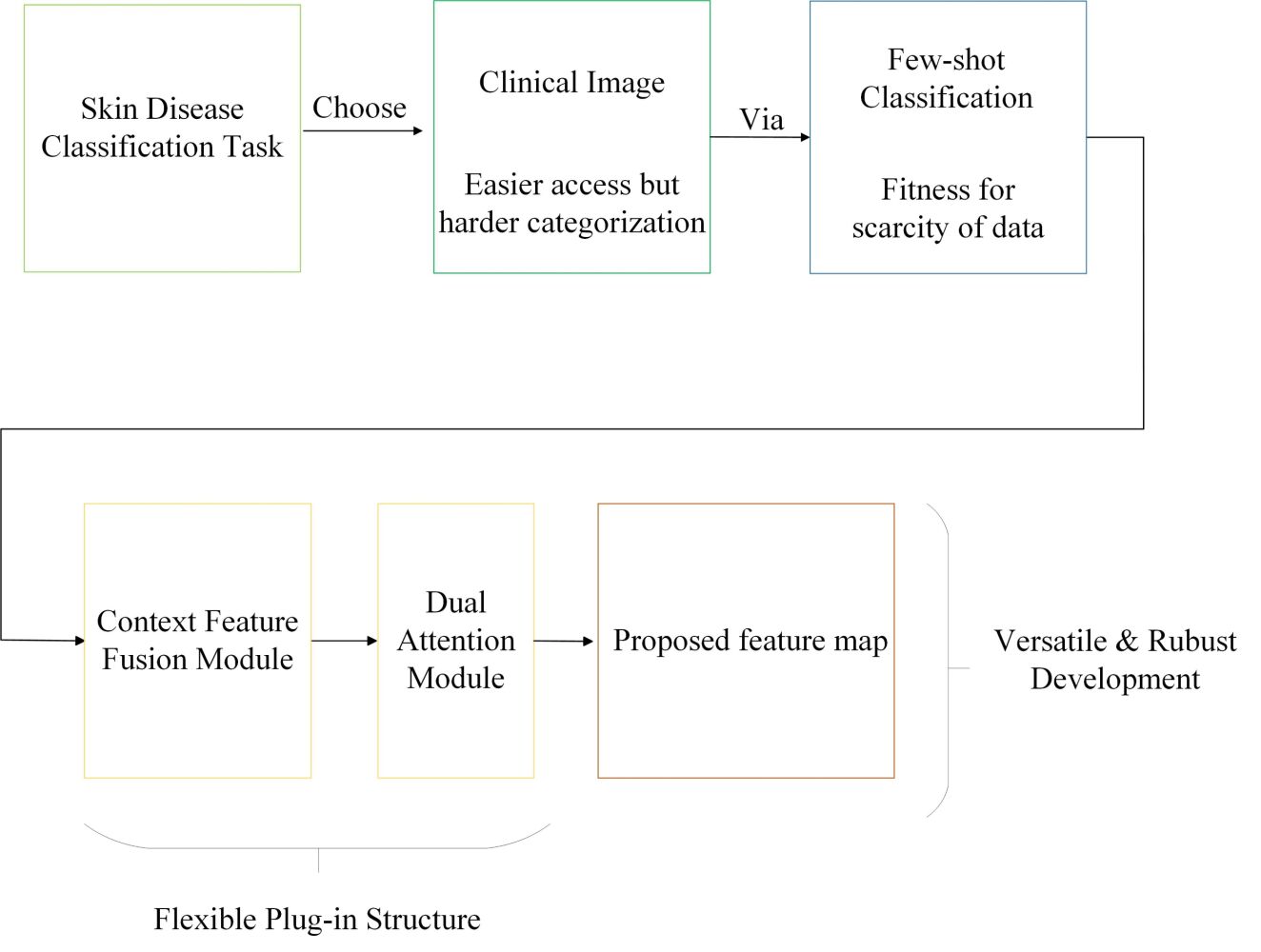
Introduction
Skin disease is one of the most common diseases and can affect people of all ages and races.1 There are many problems that skin diseases can cause to patients, including itching and bleeding, which can seriously affect their quality of life or even cause them to lose their lives. Skin diseases should be diagnosed early so that they can receive the correct treatment as soon as possible and avoid further progression. Delayed diagnosis can be attributed to the limited medical knowledge of patients and disparity in medical resources. We can solve some of these problems with the help of computer-aided diagnosis to a certain extent.
Dermoscopic images are the primary focus of early investigations using computer-aided diagnosis for skin diseases.2,3 This is due to the fact that they focus more on lesions than clinical images with uniform illumination and less noise. However, dermoscopic-based diagnosis of skin diseases has some restrictions, such as high costs and low convenience. In recent years, some researchers have begun to pay more attention to clinical images.4,5 Differences between clinical images and other images are demonstrated in Figure 1.
In recent years, the fields of feature learning and object recognition have experienced tremendous growth in the use of convolution neural networks (CNNs). According to numerous studies from ImageNet’s large-scale visual recognition challenge, the most sophisticated CNN has outperformed humans on object-classification tasks.6–8 Due to its exceptional performance over traditional methods, deep CNN-based learning is also frequently utilized in skin disease classification,9,10 lesion localization, and segmentation tasks,11–14 A majority of these tasks showed a high standard of accuracy, making automatic skin disease screening achievable.
Owing to the great amount of resources required for building a large benchmark with professional dermatologists, we believe that few-shot learning (FSL) classification would be helpful in solving the problem of scarcity of annotated data.15 It aims to accommodate novel classes unseen during training using only a few examples during testing. This is unlike a pretrained fine-tuning model, in which the classifier uses a previously learned representation and tunes its parameters to maximize the accuracy of new data. The problem with pretrained fine-tuning is that the classifier is most likely to overfit the new data when fewer examples are provided.
State of the Art
Currently, FSL methods are used frequently in early skin disease–screening application tasks such as IoMT16 and mobile diagnosis platform,17 FSL methods proven to have satisfactory performance over long-tailed and sample-classification tasks like skin disease classification. Several improved skin lesion–classification methods based on FSL have also been introduced, containing multiperspective improvements on vanilla methods. Liu et al introduced an embedding-based comparison method improved from RelationNet,18 Zhu et al proposed a novel loss-function calculation named query-relative loss over meta-learning,19 Li et al and Prabhu et al raised a similar issue of focusing on clustering methods using category judgment in feature maps,20,21 and Mahajan et al proposed an enhanced convolution strategy, ie, group-equivariant convolutions (G-convolutions) to alleviate classification error brought about by data augmentation, such as rotations and reflections.22 However, these studies were limited to nonuniversal approaches or few disease classes.
In this work, we propose a multiscale feature-fusion FSL classification approach to skin disease classification, CDD-Net (Context Feature Fusion and Dual Attention Dermatology Net), based on the special texture characteristics of clinical skin images. CDD-Net focuses on feature-level improvement, with the application of the developed extraction and fusion strategy, which makes it possible to utilize almost all the current FSL methods, rather than improvement only in specific tools, such as metric learning–based methods16,20,21 or meta-learning–based methods.18,19
To test our method, we built a large skin disease dataset, Derm104, with 104 classes of skin lesions, which provides a large benchmark for skin disease FSL classification. It merges 1781 self-collected images from a public skin disease atlas website (www.med126.com/pf) and 2702 images from the public dataset SD-198,23 for a total of 4483 images. For a more stable verification result and to ensure adequate generalization, we ensured that there were at least 20 images per category. Our contributions are as follows.
- We built a clinical skin disease dataset, Derm104, with 104 classes of skin lesion, providing a reliable benchmark for our research.
- We propose CDD-Net as a universal feature-level plug-in module in clinical skin image FSL classification fitting the special texture characteristics of skin lesions, which makes it an improvement on almost all the current FSL methods, with accuracy improvement of up to 9.14 percentage points. This approach provides a solving pattern for the scarcity of annotated data problems in clinical skin disease diagnosis.
Related Work and Materials
FSL Methods
In contrast to traditional classification methods, FSL aims to classify instances from undiscovered classes with a small number of labeled samples in each class.15 Ravi and Larochelle used an LSTM-based meta-learner that captures both short-term knowledge that is specific to a task and long-term knowledge that is shared across all tasks.24 Using the prototypes for each class, ProtoNet performs classification using a representation-based metric space.25 The use of FSL in the context of meta-learning has also been investigated by Finn et al,26 wherein they presented an algorithm for fast adaptation of networks on versatile tasks and showed their effectiveness in one-shot learning tasks. Vinyals et al proposed a network known as matching networks that learns the mapping between a small labeled support set and an unlabeled example.27 The training process is based on the idea that testing and training settings should be identical. Finn et al continued to investigate meta-learning mixed with imitation learning from a visual demonstration challenge of one-shot learning for a robot.28 Meta-learning and transfer learning were integrated by Sun et al to develop a hard-task meta-batch scheme, an effective learning curriculum that increases convergence and accuracy.29
Typically, each few-shot classification task T□ corresponds to every training episode in one epoch, and would be randomly selected from a sample dataset, and samples in task T□ would be divided into support set Ts and a query set Tq. If the support set Ts contains N classes with K samples in each class, the few-shot classification task is called N-way K-shot. The query set Tq contains the samples from the same classes with the support set Ts. Formally, a few-shot task can be defined as T=(Ts, Tq), where  and
and  . Given the support set Ts, our goal is to classify and assign the samples in the query set Tq correctly to one of the N classes. This kind of training mechanism based on task training is called metaparadigm or episodic paradigm. Figure 2 shows how episodic training works in skin disease classification.
. Given the support set Ts, our goal is to classify and assign the samples in the query set Tq correctly to one of the N classes. This kind of training mechanism based on task training is called metaparadigm or episodic paradigm. Figure 2 shows how episodic training works in skin disease classification.
There are three types of FSL classification:, pretrained fine-tuning–based methods, meta-learning–based methods, and metric learning–based methods.25–34 In pretrained fine-tuning, a generic training task is used rather than episodic training in the training stage. The entire base class Nbase is trained to train a classifier:
where  is a parameter of the embedding backbone and the classifier. In the testing stage, several episodic training tasks T were built and sampled from the domain of the novel class Nnovel. The support set Ts will fine-tune the trained
is a parameter of the embedding backbone and the classifier. In the testing stage, several episodic training tasks T were built and sampled from the domain of the novel class Nnovel. The support set Ts will fine-tune the trained  , and the query set Tq is tested in the classifier.
, and the query set Tq is tested in the classifier.
In meta-learning–based methods, episodic training is employed in both the training stage and testing stage. A meta-learner is added and the network training is processed in a two-step optimization between the base learner and meta-learner. The first step is called base learning or inner loop, which optimizes the base learner with the support set Ts, as in MAML,26 and the gradient is updated:
where  and m are the learning rate and the iteration number in the inner loop, respectively. The second step is meta-fine-tuning or outer loop, which optimizes the meta-learner with query set Tq, and a similar gradient is updated:
and m are the learning rate and the iteration number in the inner loop, respectively. The second step is meta-fine-tuning or outer loop, which optimizes the meta-learner with query set Tq, and a similar gradient is updated:
The parameter of the model is truly updated over the previous parameter  rather than
rather than  by using the query set Tq. In this way, the meta-learner is expected to learn a type of across-task meta-knowledge that can be used for fast adaptation to novel tasks.
by using the query set Tq. In this way, the meta-learner is expected to learn a type of across-task meta-knowledge that can be used for fast adaptation to novel tasks.
In metric learning–based methods, episodic training is employed in the training stage, but not in the testing stage. Instead, the testing stage will directly compare the similarities between the samples in the support set Ts and the query set Tq, as part of a single feed-forward pass mechanism. Every sample is encoded into the latent representation space, as in ProtoNet,25 and samples are encoded as prototype  , where K denotes K-shot in the support set. The prototype of the query sample XQ computes the distance with Ci, and the predicted posterior probability distributions are:
, where K denotes K-shot in the support set. The prototype of the query sample XQ computes the distance with Ci, and the predicted posterior probability distributions are:
where D (.) denotes the Euclidean distance between the prototypes. In the training stage, the standard cross-entropy loss was employed to train the entire model. In addition, during the testing stage, the nearest-neighbor classifier (1–NN) can be conveniently used for prediction.
Dataset
In order to support the application of computer-aided diagnosis in dermatology, several public datasets of skin diseases were created as a benchmark for measuring the effectiveness of automatic diagnosis. The Dermofit dataset is provided by researchers at the University of Edinburgh in Scotland.35 This dataset is of high quality and is widely used by researchers, but it is not freely available. Dermofit includes 10 different types of skin diseases, but there are only roughly 1300 photos in total. The International Skin Imaging Collaborative has collected a large dataset of publicly accessible dermoscopic images,36 which was released in 2016 for a publicly available benchmark competition for dermoscopic image analysis. MoleMap is a skin dataset consisting of three cancer categories and 22 benign categories.37 The cancer categories are melanoma, basal cell carcinoma, and squamous cell carcinoma. Each lesion image consists of a dermoscopic image and a clinical image that was captured 10 cm from the lesion. The Asan dataset was obtained from the Dermatology Department of Asan Hospital.38 It contains 12 dermatological disease categories within Asian subjects. A collection comprising 1276 of these photos is available for downloading for research purposes. SD-198 is a publicly available dataset of clinical skin images collected by Sun et al. It comprises 6584 images range in size, color, shape, and structure across 198 categories.23
All of the abovementioned datasets have their own disadvantages in clinical skin disease classification. The International Skin Imaging Collaborative dataset mainly contains dermoscopic images, which are costlier and more difficult to obtain from clinical images.36 The Dermofit, MoleMap, and Asan datasets contain fewer categories (<25) and do not cover enough rare categories.35,37,38 Compared with other datasets, SD-198 is a more proper dataset,23 but more than half of its collected categories contain fewer than 20 images, which may cause overfitting during the training of a deep-learning network. Another problem is that some major species and subspecies are not classified accurately, such as general psoriasis, pustular psoriasis, and nail psoriasis, which may cause category confusion.
We present a new clinical skin disease dataset as a benchmark for clinical skin disease classification, Derm104, which covers sufficient rare diseases and contains easily obtainable clinical images. Derm104 has high diversity, multiple categories, wide coverage, and reliable samples and benchmark experiments can effectively evaluate the reliability of various FSL methods in clinical skin disease classification.
- Annotation and Scale: Derm104 contains 4483 clinical images covering 104 types of skin disease and is merged from two parts. The first part comprised 1781 images, all of which were collected by professional dermatologists from the public dermatology dataset on www.med.126.com/pf. The second part contains 2702 images from the public dataset SD-198. Both parts of the dataset are public and freely available to researchers, and the patients involved in the database have given approval for their images to be used. To prevent ambiguity and ensure the accuracy of classification between subspecies, we chose some of the images from SD-198, and every category ground truth was verified by professional dermatologists. Images per class vary from 20 to 141 to ensure variance stability.
- Diversity: All images are from a real scene with variance in color, exposure, illumination, and level of detail collected in a variety of environments. Also, skin lesion images include: eczema, psoriasis, acne, lichens, prurigo, alopecia areata, urticaria, scabies, nervus, erythema, pityriasis, vercurra, bacterial skin diseases, and melanoma skin cancer, covered more than 90% of the incidence of all skin diseases.
- Fine Granularity: Our dataset also focuses on fine-grained disease classes, such as many subspecies that have tiny interclass variance and are difficult to classify. For example, erythema, pityriasis, and lichen diseases can be classified into many subcategories that are difficult to distinguish. Few subspecies are included in existing datasets,35–38 so Derm104 focuses more on this. Some of our fine-grained images are shown in Figure 3.
Methods
The classification of clinical skin lesions emphasizes lesion location and examination of textural features compared with natural scene images. For example, locations such as the cheek, nose, or eye influence the determination of facial skin diseases, and texture features such as line shape and large flake area cannot be easily focused on, rather than dot or block shapes. However, normal classification methods may not work efficiently. In order to extract specific deep features in skin lesions, we propose CDD-Net as an enhanced solution for clinical skin disease classification. Current FSL methods for skin disease classification are limited to individual approaches,18–22 but CDD-Net has highly versatile performance that can be used as a plug-in module in both metric learning–based, pretrained fine-tuning–based and meta-learning–based FSL methods.
CDD-Net is composed of a context feature–fusion module and a dual-attention module for different stages of feature processing. The context feature–fusion module localizes the discriminatory texture details of skin lesions by integrating features from different layers. The dual-attention module highlights discriminative regions from channel-wise and pixel-wise representations based on weight vectors and restrains the contributions of irrelevant areas. In the following section, we describe the entire framework of the proposed CDD-Net. The overall flowchart is given in Figure 4.
Context Feature–Fusion Module
In recent years, the idea of feature fusion has been applied in many visual tasks. Specifically, Long et al combined deep semantic features with shallow appearance features to produce accurate and detailed segmentations.39 Manzoor et al effectively fused deep features extracted from different convolution layers for high-resolution dermoscopic skin image classification.40 In this paper, we introduce a context feature–fusion mechanism to exploit the strong complementary and correlated information among different hierarchical layers for clinical skin image classification.
Considering that different layers may have feature maps of different sizes, we use the bottleneck module to ensure that they have the same spectral dimensionality before feature fusion. Such an operation is applicable to convolution layer-based neural networks, and thus we can utilize our module within CNN networks such as ResNet and others. For instance, in ResNet12, we assume that F1, F2, F3, and F4 refer to the outputs from low to higher-level layers that have 64, 160, 320, and 640 feature maps, respectively. Then, we can use 640 kernels with of size 1×1 to convolute them. With such a convolution operation, the number of feature maps from F1 to F4 becomes 640. Finally, feature fusion can be performed easily using element-wise summation. The entire process can be represented as:
where FZ represents the fused features, FL represents features from different convolution layers, and BL is a bottleneck module corresponding to FL. Conversely, FZ can also be represented as the result of sample image xi processed through our feature extractor hθ,µ, where hθ,µ is combined by convolution network gθ and feature-fusion module fµ:
Finally, we use pooling as the global averaging function. The entire context feature–fusion module is illustrated in Figure 5.
Dual-Attention Module
Introducing an attention mechanism to complete the adaptive weight adjustment is helpful in highlighting the representative discriminative regions and restraining the contributions of irrelevant backgrounds. Ma et al used a squeeze-and-excitation module to enhance channel weight on new feature maps via pooling and full connection layers, elevating segmentation efficiency for brain tumor imags.41 Space weight focuses on the location of the area of interest. Wei et al proposed a spatial pyramid pooling module for lesion segmentation of dermoscopic images.42
We propose a dual-attention mechanism to enhance the ability to exploit features of interest based on sensitivity scores. For each episode of the support set and query set, we define the channel-wise and pixel-wise sensitivity score to quantify the representativeness. Prior to explaining scores, we first define feature maps F of the support and query instance as follows:
where  is the j-th instance of the i-th class in the support set, Xq is the query instance, and hθ,µ is a feature extractor supported by the proposed context-feature fusion in (Equation 6). Specifically, we utilize a prototype as the representative of each class in the support set.25 The prototype is defined as:
is the j-th instance of the i-th class in the support set, Xq is the query instance, and hθ,µ is a feature extractor supported by the proposed context-feature fusion in (Equation 6). Specifically, we utilize a prototype as the representative of each class in the support set.25 The prototype is defined as:
where k is the instance size for each support class (ie, k-shot). Based on this,  and
and  mean the c-th channel and x-th pixel of the prototype feature map
mean the c-th channel and x-th pixel of the prototype feature map  , respectively, and we compute the mean features in channel and spatial dimensions to represent the standard condition in each class:
, respectively, and we compute the mean features in channel and spatial dimensions to represent the standard condition in each class:
Furthermore, we obtain the channel-wise and pixel-wise sensitivity scores for each channel and pixel. Note that we compute scores in the feature maps of support images rather than prototypes, where  indicates the feature map on the j-th instance of the i-th class in the support set. Similarly to what we have calculated on the aforementioned prototype feature,
indicates the feature map on the j-th instance of the i-th class in the support set. Similarly to what we have calculated on the aforementioned prototype feature,  and
and  refer to the c-th channel and x-th pixel of
refer to the c-th channel and x-th pixel of  , respectively:
, respectively:
Intuitively, sensitive scores represent the discriminative information between the mean standard and region of interest, but cannot finally determine how much attention should be paid to this. In order to choose efficient weights and improve the generalization, we introduce the fully connected block (FCB) to realign the weight vectors. The architecture of the FCB is presented in Table 1.
 |
Table 1 Architecture of the fully connected block.Fully Connnection Block |
Notes: S, input weight that would be realigned. When FCB is required for adaption of channel-wise depiction, S is equal to channel size C, while FCB requires pixel-wise depiction and S is equal to layer size H×W.
The sensitivity scores in a single-feature map can be combined into weight vectors  and
and  , and they would be transformed through a fully connected block corresponding to the discriminative information from the perspectives of channel and pixel, respectively, and processed thus:
, and they would be transformed through a fully connected block corresponding to the discriminative information from the perspectives of channel and pixel, respectively, and processed thus:
where H represents FCBs that adapt to the different weight vectors. In the episode-training stage, we add random noise between −0.2 and 0.2 to the weight vector vi,j to prevent overfitting. Upon obtaining channel- and pixel-discrimination weight vectors, we scale them with the original feature map correspondingly. Finally, we construct a linear combination with a balancing hyperparameter α:
In a similar vein, we can also obtain the processed feature map  from the query image. The difference is that we use its own feature map rather than the prototype in the support set. The entire dual-attention module is shown in Figure 6.
from the query image. The difference is that we use its own feature map rather than the prototype in the support set. The entire dual-attention module is shown in Figure 6.
 |
Figure 6 Schematic illustration of dual-attention module. The different colors in the cubes indicate different sensitive scores in channels and pixels. In addition, calculations of sensitive scores for individual feature maps in the support set and query set, channel-wise and pixel-wise, are independent of one another. The final combination from the perspectives proposed is dependent on hyperparameter α ∈ [0,1] (Equation 12), which is omitted in the illustration. |
Results
In this section, we evaluate CDD-Net on the proposed Derm104 benchmark. Derm104 consists of 4483 clinical images covering 104 types of skin diseases. Derm104 has detailed and reliable labels, enough categories, high diversity, and fine granularity, and thus we propose it as our benchmark for clinical skin disease FSL classification. In sum, 62 categories were set as the training set, with 21 categories as the validation set and another 21 categories as the testing set.
We adopted common protocols from recent FSL research on skin disease.18–22 The entire training procedure was completed on the GPU of an Nvidia GeForce 1080 Ti. The input image size was set to 84×84×3, and lightweight backbones of Conv64F and ResNet12 were used. ResNet12 produced a feature map with an output size of 640×5×5, whereas Conv64F provided a size of 64×5×5. Prior to inputting the photos into the convolution network, common data-augmentation methods, such as center crop and horizontal flip, were employed. Note that skin lesion classification is color-sensitive, so color jitter is not used.43 Parameter α (Equation 12) was fixed at 0.5.
Following common FSL methods,15,24,27 we set five-way one-shot and five-way five-shot for the episode-training paradigm and 10 query images per new class for result evaluation. Episodes of training, validation, and testing were set to 100 per epoch, and the entire learning procedure consisted of 100 epochs. We set a learning rate of 1–3 for the Adam optimizer. As for some approaches that require another learning rate in their inner circle, we set rates of 1–3 for ANIL and 1–2 for baseline++ and MAML. In the fully convoluted block of the dual-attention module, we set a rate of 1–4. All these learning rates were step-scheduled to 0.1 after 70 epochs.
To verify the high adaptability of CDD-Net, we applied it to various FSL methods from different approaches: the pretrained fine-tuning–based methods baseline++ and RFS-Distill, meta-learning–based methods MAML, ANIL, metric learning–based methods ProtoNet, DN4, EPNet, and Neg-Cosine.25–27,30–34The classification accuracy was within the 95% confidence interval. Tables 2 and 3 show the performance of our proposed CDD-Net on five-way one-shot and five-way five-shot clinical skin disease classification, respectively.
 |
Table 2 Performance of CDD-Net on five-way one-shot classification with comparison of various methods |
 |
Table 3 Performance of CDD-Net on five-way five-shot classification with comparison of various methods |
As shown in Tables 2 and 3, we clearly validated the impressive performance, generalization ability, and robustness of the proposed CDD-Net. It is versatile for pretrained fine-tuning–based, meta-learning–based, and metric learning–based FSL methods, with accuracy improvement of up to 9.14 percentage points. Since CDD-Net has better performance on metric learning–based approaches, we conducted an ablation study on ProtoNet, DN4, EPNet, and Neg-Cosine with the modified CDD-Net and report the efficiency of its submodules.
We additionally explored the effects of the dual-attention module in CDD-Net. In Table 4, the first row shows the results without any modules and the third row shows the accuracy after adding dual attention. We found that perceiving the relative location of skin lesion and screening task-related regions is as important as emphasizing the channels that extract key features. A single attention model is not sufficiently effective, and the second row in Table 4 shows the influence of using only a channel-wise sensitivity score in the classification.
 |
Table 4 Performance of different attention-selection strategies in dual-attention module |
Furthermore, we studied the fusion strategy in the context feature–fusion module. The first row in Table 5 shows the results without any feature-fusion operations, as well as the third row in Table 4. The results of concatenative and additive fusion are shown in the second and third rows, respectively. We suppose that the concatenation of different layers causes the channel dimension to overinflate (up to 2560 in ResNet12), which may cause overfitting and dimension disaster. The bottleneck convolution in the last layer would lose critical gradient information. Instead, additive fusion using average pooling is an easy and reliable approach.
 |
Table 5 Performance of different fusion strategies in context feature–fusion module |
 |
Table 6 Comparison of different methods on several different evaluation metrics |
In order to obtain a better measure of the detailed improvement of CDD-Net, for each class of clinical skin disease classification, we add a comparison experiment between DN4 with CDD-Net and several vanilla networks relying on ResNet12 embedding. We demonstrate the result of final epoch running on the testing set comprising 21 categories. This epoch contains 100 episodes of five-way five-shot classification, ie, 50 query images in five ways per episode, for a total of 5000 queries randomly selected from the testing set. We have chosen several methods that perform better than the aforementioned results, including the metric learning method DN4, EPNet, and the pretrained fine-tuning–based method RFS-Distll for further demonstration. The comparison of confusion matrices for query images between vanilla DN4 and CDD-Net is illustrated in Figures 7 and 8.
 |
Figure 7 Confusion matrix with vanilla DN4. |
 |
Figure 8 Confusion matrix with CDD-Net. |
To evaluate the performance of our proposed method, several evaluation metrics are employed: accuracy  , precision
, precision  , recall
, recall  , and F1 score
, and F1 score  . These evaluation metrics are defined based on confusion matrix in which TP, TN, FP, and FN refer to true positive (both real and predicted classes are true), true negative (both real and predicted classes are false), false positive (the actual class of the data is false, while the predicted one is true), and false negative (the actual class of the data is true, while the predicted one is false), respectively. Accuracy is the number of correct predictions over the total number of predictions, also called the classification rate. The number of predicted positive classes that belong to the actual positive class is calculated by recision, while recall determines the true-positive rate. F1 score measures the testing result according to precision and recall. The comparison of CDD-Net with vanilla DN4, EPNet, and RFS-Distill is illustrated in Figures 9–11.
. These evaluation metrics are defined based on confusion matrix in which TP, TN, FP, and FN refer to true positive (both real and predicted classes are true), true negative (both real and predicted classes are false), false positive (the actual class of the data is false, while the predicted one is true), and false negative (the actual class of the data is true, while the predicted one is false), respectively. Accuracy is the number of correct predictions over the total number of predictions, also called the classification rate. The number of predicted positive classes that belong to the actual positive class is calculated by recision, while recall determines the true-positive rate. F1 score measures the testing result according to precision and recall. The comparison of CDD-Net with vanilla DN4, EPNet, and RFS-Distill is illustrated in Figures 9–11.
 |
Figure 9 Comparison of precision in CDD-Net, vanilla DN4, EPNet, and RFS-Distill. |
 |
Figure 10 Comparison of recall in CDD-Net, vanilla DN4, EPNet, and RFS-Distill. |
 |
Figure 11 Comparison of F1-Score in CDD-Net, vanilla DN4, EPNet, and RFS-Distill. |
The comparison of complex matrix and the precision, recall, and F1 scores focuses on the detailed performance of different classification approaches on different disease categories. The preceding experiment shows that our proposed CDD-Net performs best in most of the testing categories, especially in Cl (cellulitis) and Hn (halo nevus), with a lead nearing 10 percentage points over DN4, but it does not show enough advantage in Dp (dilated pore of Winer), Ns (nevus sebaceous of Jadassohn), or Oc (Onychomycosis).
Top-1 accuracy is the accuracy of the top-ranked category that matches the actual results, while Top-5 accuracy is the accuracy of the top five categories containing actual results. Macro-averaging and micro-averaging methods are also widely used in judging multiclassification tasks. Macro-averaging counts the indicator values for each class and then averages them arithmetically across all classes:  ,
,  ,
,  . Micro-averaging calculates the corresponding metrics based on a global confusion matrix:
. Micro-averaging calculates the corresponding metrics based on a global confusion matrix:  . The comparison of different methods on several evaluation metrics is demonstrated in Table 6.
. The comparison of different methods on several evaluation metrics is demonstrated in Table 6.
Discussion
Compared with traditional classification methods, FSL classification performs better in accommodating unseen novel classes15 and has demonstrated great suitability and superiority in the field of clinical dermatology diagnostics,16,17 since skin disease–classification tasks have have large variety and few samples. The classification accuracy was within the 95% confidence interval, and shows our CDD-Net is a versatile and robust development for clinical skin disease FSL classification. Despite FSL approaches using different training paradigms, such as pretrained fine-tuning–based, meta-learning–based, and metric learning–based methods, CDD-Net performs more efficiently and is stable. Each submodule in the multifusion strategy has also been shown to be efficient, proving that our improved model in this work is feasible and correct.
Most approaches perform better with more complex backbones, except for meta-learning methods, which are unstable in sensitive networks. Based on the results in Tables 2 and 3, CDD-Net shows better performance on metric learning–based methods than others, especially for the five-way five-shot ResNet12-embedded network: a 4.87 percentage-point improvement versus the pretrained fine-tuning–based method RFS-Distill. We suppose that the reason is that metric learning–based methods have points on the comparison of feature maps between the support set and query set, and our optimization of feature maps fits better to this. However, pretrained fine-tuning–based and meta-learning–based approaches have more dependence on the optimization of the training mechanism. Pretrained fine-tuning–based methods rely more on prior knowledge in the base classes, whereas meta-learning–based methods rely more on justified meta-learner iterations.
It is worth noting that CDD-Net’s performance was unsatisfactory when compared to some approaches, including baseline++. We suppose that this is due to our sensitivity scores calculated in the dual-attention module being based on Euclidean distance, whereas baseline++ uses the cosine distance instead of the linear layer as its part of the classifier. Cosine distance focuses more on relative direction, but is not sensitive to absolute distance, and thus the classifier cannot take sufficient advantage of our provided template. The result for Neg-Cosine also shows less accuracy improvement than that over other methods. CDD-Net performance was more robust in five-shot implementations, indicating its reliability with more samples to learn and feedback than standard methods. CDD-Net showed improvement in versatility of 0.93 to 9.14 percentage points over vanilla approaches, and the best results were for metric learning–based methods.
We chose some skin lesion images from our dataset for Grad-CAM visualization (Figure 12).44 Three types of representative result image were randomly selected: benign keratosis, tinea corporis, and junction nevus. To show the tendency of the judgment in the neural network, Grad-CAM results for the last layer in the network are demonstrated. Since the ablation results on ProtoNet in five-way five-shot mode showed better improvement, we choose these Grad-CAM results for a more intuitive demonstration. We found that the network’s “attention” to the actual lesion becomes more accurate with the improvement in the various modules of the CDD-NET. Before the dual-attention module was applied,the tendency of the network to locate a lesion was not sufficiently obvious, and there will be cases in which the zone of interest is confusing and mistaken.
Due to the difficulty of obtaining a larger number (around 40 or 50 images per category compared with 600 samples per category in the natural landscape and object image dataset ImageNet) of images for clinically rare skin diseases, we can only ensure a minimum of 20 images per category in Derm104, so this might be a limitation whereby trained neural network generalization is not good enough for certain disease categories. We hope that those in the field of dermatology are able to collect and collate more images of skin diseases of clearer quality, especially in rare disease categories, with the development of automated health care.
Conclusion
In this paper, we introduce CDD-Net, a versatile plug-in module for clinical skin disease FSL classification, which performs well in different FSL approaches, including pretrained fine-tuning–based, meta-learning–based, and metric learning–based methods. CDD-Net presents a context feature–fusion module and dual-attention module to extract discriminating texture features and emphasize contributive region and channels. We also present Derm104, a new clinical skin disease dataset with significant coverage for rare diseases and a reliable annotation between primary species and subspecies, as our benchmark. CDD-Net performed better on metric learning–based methods and has an accuracy improvement of up to 9.14 percentage points. Our ablation study validated its flexible structure and the importance of each component. As a future direction, we will investigate how to locate and extract skin lesion features of different sizes and shapes on complex and various backgrounds and summarize a better extraction logic, specifically for clinical skin backgrounds.
Data Sharing
The experimental dataset combines data from two sources. The first part was obtained from the public dermatology atlas website www.med126.com/pf. The second part is from the available dataset SD-198, collected by Sun et al.23
Ethics and Consent
Experimental data were selected from SD-198 and www.med126.com/pf. These two datasets are public and freely available for researchers, They were assembled in accordance with the 1964 Declaration of Helsinki and later amendments or comparable ethical standards. The patients whose records are in the database have given approval for their use. Users can download relevant data free for research and publish relevant articles. Our study is based on open-source retrospective data that had already been collected by other researchers. No new samples were collected from any patients, so there are no ethical issues or conflicts of interest.
SD-198 is a public skin disease dataset provided by Sun et al23 that contains 6783 images of various skin lesions. This dataset has already led to improvements in dermatology research and is publicly available for anyone in the research community to use. The website www.med126.com/pf is a medical forum for students and researchers for study and discussion. Clinical skin disease images in many classes are demonstrated for study or research and are also freely available to download. Our research is free from ethical issues, based on the “Approaches to ethical review of life sciences and medical research” in China, as shown in the following paragraph.
According to items 1 and 2 of Article 32 of the Measures for Ethical Review of Life Science and Medical Research Involving Human Subjects, which was reviewed by the National Science and Technology Ethics Committee, approved by the State Council of China, and jointly promulgated by the National Health Commission, Ministry of Education, Ministry of Science and Technology and the State Administration of Traditional Chinese Medicine on February 18, 2023, stipulates the following: 1. The research is conducted using legally obtained public data or data generated through observation and does not interfere with public behavior. 2. Use of anonymized data for research. For more information on China’s guidelines on research ethics and governance, please see https://www.gov.cn/zhengce/zhengceku/2023-02/28/content_5743658.htm.
Funding
This work was not supported by any funding source.
Disclosure
The authors declare no conflict of interest.
References
1. Hay RJ, Johns NE, Williams HC, et al. The global burden of skin disease in 2010: an analysis of the prevalence and impact of skin conditions. J Invest Dermatol. 2014;134(6):1527–1534. doi:10.1038/jid.2013.446
2. Gonzalez-Castro V, Debayle J, Wazaefi Y, et al. Automatic classification of skin lesions using color mathematical morphology-based texture descriptors.
3. Badano A, Revie C, Casertano A, et al. Consistency and standardization of color in medical imaging: a consensus report. J Dig Imag. 2015;28:41–52. doi:10.1007/s10278-014-9721-0
4. Jafari MH, Karimi N, Nasr-Esfahani E, et al. Skin lesion segmentation in clinical images using deep learning.
5. Yang J, Sun X, Liang J, Rosin PL. Clinical skin lesion diagnosis using representations inspired by dermatologist criteria.
6. Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift.
7. Szegedy C, Liu W, Jia Y, et al. Going deeper with convolutions.
8. Russakovsky O, Deng J, Su H, et al. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–252. doi:10.1007/s11263-015-0816-y
9. Junayed MS, Islam MB, Jeny AA, Sadeghzadeh A, Biswas T, Shah AS. ScarNet: development and validation of a novel deep CNN model for acne scar classification with a new dataset. IEEE Access. 2021;10:1245–1258. doi:10.1109/ACCESS.2021.3138021
10. X-y Z, Wu X, F-f L, et al. The application of deep learning in the risk grading of skin tumors for patients using clinical images. J Med Syst. 2019;43:1–7.
11. Xie Y, Zhang J, Xia Y, Shen C. A mutual bootstrapping model for automated skin lesion segmentation and classification. IEEE Transac Med Imag. 2020;39(7):2482–2493. doi:10.1109/TMI.2020.2972964
12. Khan MA, Zhang Y-D, Sharif M, Akram T. Pixels to classes: intelligent learning framework for multiclass skin lesion localization and classification. Comput Electr Eng. 2021;90:106956.
13. Balaji VR, Suganthi ST, Rajadevi R, Kumar VK, Balaji BS, Pandiyan S. Skin disease detection and segmentation using dynamic graph cut algorithm and classification through Naive Bayes classifier. Measurement. 2020;163:107922. doi:10.1016/j.measurement.2020.107922
14. Roy K, Chaudhuri SS, Ghosh S, Dutta SK, Chakraborty P, Sarkar R. Skin Disease detection based on different Segmentation Techniques.
15. Lake B, Salakhutdinov R, Gross J, Tenenbaum J. One shot learning of simple visual concepts.
16. Xiao J, Xu H, Fang D, Cheng C, Gao H. Boosting and rectifying few-shot learning prototype network for skin lesion classification based on the internet of medical things. Wireless Networks. 2021;2021:1–15.
17. Lee K, Cavalcanti TC, Kim S, et al. Multi-task and few-shot learning-based fully automatic deep learning platform for mobile diagnosis of skin diseases. IEEE J Biomed Health Inform. 2022;27(1):176–187. doi:10.1109/JBHI.2022.3193685
18. Liu X-J, K-l L, H-y L, W-h W, Z-y C. Few-shot learning for skin lesion image classification. Multimedia Tools Appl. 2022;81(4):4979–4990. doi:10.1007/s11042-021-11472-0
19. Zhu W, Liao H, Li W, Li W, Luo J. Alleviating the incompatibility between cross entropy loss and episode training for few-shot skin disease classification.
20. Shuhan LI, Li X, Xu X, Cheng K-T. Sub-cluster-aware network for few-shot skin disease classification. arXiv preprint arXiv. 2022;2022:1.
21. Prabhu V, Kannan A, Ravuri M, Chaplain M, Sontag D, Amatriain X. Few-shot learning for dermatological disease diagnosis.
22. Mahajan K, Sharma M, Vig L. Meta-dermdiagnosis: few-shot skin disease identification using meta-learning.
23. Sun X, Yang J, Sun M, Wang K. A benchmark for automatic visual classification of clinical skin disease images.
24. Ravi S, Larochelle H. Optimization as a model for few-shot learning.
25. Snell J, Swersky K, Zemel R. Prototypical networks for few-shot learning. Advan Neural Inform Process Sys. 2017;2017:30.
26. Finn C, Abbeel P, Levine S. Model-agnostic meta-learning for fast adaptation of deep networks.
27. Raghu A, Raghu M, Bengio S, Vinyals O. Rapid learning or feature reuse? Towards understanding the effectiveness of maml. arXiv preprint arXiv. 2019;2019:1.
28. Finn C, Yu T, Zhang T, Abbeel P, Levine S. One-shot visual imitation learning via meta-learning.
29. Sun Q, Liu Y, Chua T-S, Schiele B. Meta-transfer learning for few-shot learning.
30. Chen W-Y, Liu Y-C, Kira Z, Wang Y-CF, Huang J-B. A closer look at few-shot classification. arXiv preprint arXiv. 2019;2019:1.
31. Tian Y, Wang Y, Krishnan D, Tenenbaum JB, Isola P. Rethinking few-shot image classification: a good embedding is all you need?
32. Li W, Wang L, Xu J, Huo J, Gao Y, Luo J. Revisiting local descriptor based image-to-class measure for few-shot learning.
33. Rodrguez P, Laradji I, Drouin A, Lacoste A. Embedding propagation: smoother manifold for few-shot classification.
34. Liu B, Cao Y, Lin Y, et al. Negative margin matters: understanding margin in few-shot classification.
35. Ballerini L, Fisher RB, Aldridge B, Rees J. A color and texture based hierarchical K-NN approach to the classification of non-melanoma skin lesions. Color Med Image Anal. 2013;2013:63–86.
36. Codella NCF, Gutman D, Celebi ME, et al. Skin lesion analysis toward melanoma detection: a challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (ISIC).
37. Yi X, Walia E, Babyn P. Unsupervised and semi-supervised learning with categorical generative adversarial networks assisted by Wasserstein distance for dermoscopy image classification. arXiv preprint arXiv. 2018;2018:1.
38. Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol. 2018;138(7):1529–1538. doi:10.1016/j.jid.2018.01.028
39. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation.
40. Manzoor K, Majeed F, Siddique A, et al. A lightweight approach for skin lesion detection through optimal features fusion. Comput Mater Continua. 2022;70(1):1617–1630. doi:10.32604/cmc.2022.018621
41. Ma S, Tang J, Guo F. Multi-task deep supervision on attention R2U-net for brain tumor segmentation. Front Oncol. 2021;11:704850. doi:10.3389/fonc.2021.704850
42. Wei Z, Shi F, Song H, Ji W, Han G. Attentive boundary aware network for multi-scale skin lesion segmentation with adversarial training. Multimedia Tools Appl. 2020;79:27115–27136. doi:10.1007/s11042-020-09334-2
43. Kasmi R, Mokrani K. Classification of malignant melanoma and benign skin lesions: implementation of automatic ABCD rule. IET Image Process. 2016;10(6):448–455. doi:10.1049/iet-ipr.2015.0385
44. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-cam: visual explanations from deep networks via gradient-based localization.
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


















