Back to Journals » Infection and Drug Resistance » Volume 13
Virulence-Related Gene Distribution Among Shigella Isolates in Anhui, China: The Association with Antimicrobial Resistance
Authors Liu Y , Shen W, Sun Y, Lv N, Sun K, Hu L , Ye Y, Gao Y , Li J
Received 30 July 2020
Accepted for publication 26 September 2020
Published 19 October 2020 Volume 2020:13 Pages 3637—3647
DOI https://doi.org/10.2147/IDR.S274862
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Yanyan Liu,1– 3,* Weihua Shen,4,* Yating Sun,1,* Na Lv,1 Kaili Sun,5 Lifen Hu,1 Ying Ye,1– 3 Yufeng Gao,1– 3 Jiabin Li1– 3,5
1Department of Infectious Diseases, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, People’s Republic of China; 2Anhui Center for Surveillance of Bacterial Resistance, Hefei, Anhui, People’s Republic of China; 3Institute of Bacterial Resistance, Anhui Medical University, Hefei, Anhui, People’s Republic of China; 4Department of Special Clinic, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, People’s Republic of China; 5Department of Infectious Diseases, The Chaohu Affiliated Hospital of Anhui Medical University, Hefei, Anhui, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jiabin Li
Department of Infectious Diseases, The First Affiliated Hospital and the Chaohu Affiliated Hospital of Anhui Medical University, Jixi Road No. 218, Hefei 230022, Mainland China
Tel +86-551-62922713
Fax +86-551-62922281
Email [email protected]
Objective: The aim of this study was to investigate the antimicrobial resistance profiles and distribution of virulence-related genes (VRGs) among Shigella isolates in Anhui, China, and to identify the correlation between the VRGs and antimicrobial resistance.
Materials and Methods: A total of 525 non-duplicate Shigella isolates (449 S. flexneri, 68 S. sonnei, 3 S. boydii, and 5 S. dysenteriae) were collected in Anhui Province, China between September 2011 and September 2015. The antimicrobial resistance of the strains was determined by the agar dilution method according to CLSI guidelines. The presence of 16 VRGs, including ipaH, ipaA-D, ial, virB, virF, set, sen, icsA, icsB, sigA, sat, pic, and sepA, was evaluated using PCR amplification and sequencing.
Results: Shigella flexneri was the most abundant (85.5%), followed by S. sonnei (13.0%). The proportion of males with S. flexneri was higher than that of females (57% vs 43%; P< 0.0001). The most common resistance pattern was the combination of ampicillin, nalidixic acid, and tetracycline for S. flexneri (90.2%) and S. sonnei (94.1%). Resistance to ciprofloxacin and levofloxacin was more common among S. flexneri than among S. sonnei (49.7% vs .19.1%, P< 0.0001; 30.5% vs 10.3%, P=0.001, respectively). All the isolates were positive for the ipaH gene, while the set, sat, pic, and sepA genes were not detected among the S. sonnei isolates. Except for sigA and sen, resistance to chloramphenicol and ciprofloxacin was more common among VRG-positive S. flexneri than among VRG-negative S. flexneri (P< 0.05). Furthermore, resistance to ceftriaxone and ceftazidime was more frequently detected among sat- and set-positive S. flexneri than among sat- and set-negative S. flexneri (P< 0.05). However, gentamicin resistance was more prevalent among VRG-negative (ial, virF, set, sat, pic, and sepA) S. flexneri than among VRG-positive S. flexneri (P< 0.05).
Conclusion: Shigella flexneri remains the predominant species in Anhui, China, and the resistance to fluoroquinolones was more widespread among S. flexneri than among S. sonnei. Shigella flexneri strains harboring specific VRGs were associated with antimicrobial resistance. To the best of our knowledge, this is the first report of the correlation between the VRGs and antimicrobial resistance in Anhui, China.
Keywords: antimicrobial resistance, Shigella flexneri, Shigella sonnei, virulence-related gene, type III secretion system
Introduction
Shigellosis is one of the most important causes of diarrhea worldwide.1 The annual number of deaths due to shigellosis has been estimated at 1.1 million, and shigellosis-associated mortality is particularly prevalent among children in developing countries.2 Shigellosis can be caused by four Shigella species: S. dysenteriae, S. flexneri, S. boydii, and S. sonnei. Shigella sonnei is the predominant serogroup in developed countries, while S. flexneri is the main serogroup found in developing countries.3 However, the frequency of S. sonnei has recently increased in several areas that have undergone rapid socioeconomic improvements.4,5
Antimicrobial agents have been increasingly effective in alleviating the dysenteric syndrome associated with shigellosis.1 However, the effectiveness of antimicrobial agents has become limited due to the emergence of multidrug resistance (MDR).3 Antimicrobial resistance among Shigella species, including resistance to fluoroquinolones and third-generation cephalosporins, is prevalent in China.6–8 Moreover, antimicrobial resistance differs among regions.9 Consequently, it is important to understand the nature of the local antimicrobial resistance when choosing an appropriate antibiotic for the treatment of shigellosis.
The type III secretion system (T3SS) of Shigella has been reported to be associated with the pathogenesis of shigellosis, and is regulated by multiple virulence-related genes (VRGs). The differential distribution of VRGs among Shigella isolates can lead to different clinical manifestations.10,11 Invasion plasmid antigen (ipa) genes and invasion associated locus (ial) are important mediators of Shigella cell invasion in the intestinal epithelium.12 Shigella also produces distinct enterotoxins such as Shigella enterotoxin 1 (ShET-1), encoded by the set gene, and Shigella enterotoxin 2 (ShET-2), encoded by the sen gene.10 VirF and VirB (InvE) are transcriptional activators of plasmid-bourne genes that control the expression of invasion-related genes.13 The icsA gene is the major determinant of Shigella actin-based motility, and has been shown to interact with icsB through the prevention of autophagic recognition of icsA.14 Finally, the genes encoding serine protease autotransporters of Enterobacteriaceae (SPATE) are phylogenetically classified into two main classes: class 1 SPATE genes, comprising the Shigella IgA-like protease homolog (sigA) and secreted autotransporter toxin (sat); and class 2 SPATE genes, comprising Shigella extracellular protein A (sepA) and protease involved in intestinal colonization (pic).15 Overall, Shigella isolates harboring these VRGs can induce inflammation and extensive mucosal damage during intestinal infections, especially when the isolates encode more than one VRG. Furthermore, the detection of additional VRGs in Shigella spp. together with the concomitant increase in resistance against more antimicrobials suggests that there is a link between virulence and antimicrobial resistance of shigellosis.11 Hence, understanding the distribution of VRGs in Shigella isolates is useful for designing new antimicrobial therapies based on gene targets.
To the best of our knowledge, this is the first report of the VRG profiles of Shigella isolates in Anhui, China. The aim of this study was to investigate the antimicrobial resistance profiles and VRG distribution among Shigella isolates in Anhui, China, and identify the correlation between the VRGs and antimicrobial resistance to help guide the treatment of shigellosis.
Materials and Methods
Bacterial Isolates and Serotyping
A total of 525 non-duplicate Shigella isolates (449 S. flexneri, 68 S. sonnei, 3 S. boydii, and 5 S. dysenteriae) were collected from the stool samples of patients in Anhui Province, China between September 2011 and September 2015. Individual isolates were identified through standard microbiological and biochemical methods. All Shigella isolates were confirmed via the VITEK 2 Compact system (bioMérieux, Marcy l’ Étoile, France) and serotyped using commercially available antisera (Denka Seiken Co., Ltd, Tokyo, Japan). Escherichia coli ATCC 25922 was stored at the Anhui Center for the Surveillance of Bacterial Resistance (Hefei, Anhui, China). All clinical samples were part of the routine hospital laboratory procedure.
The study was conducted in accordance with the guidelines of the Declaration of Helsinki, the principles of Good Clinical Practice, and Chinese regulatory requirements, and was approved by the local Ethics Committees of the First Affiliated Hospital of Anhui Medical University (Hefei, China). All patients gave written informed consent, and parental/legal guardian informed consent was obtained for patients under the age of 18.
Antimicrobial Susceptibility Testing
Bacteria were cultured to the log-phase (approximately 1.5 × 108 CFU/mL). The minimum inhibitory concentrations (MICs) of the following 17 antimicrobial agents were determined by the agar dilution method using Mueller-Hinton agar (Oxoid Ltd, Basingstoke, UK) containing a series of twofold diluted antibiotics: ampicillin (AMP), nalidixic acid (NAL), tetracycline (TCY), chloramphenicol (CHL), trimethoprim/sulfamethoxazole (SXT), cefotaxime (CTX), ceftriaxone (CRO), ceftazidime (CAZ), cefepime (FEP), cefoxitin (FOX), ciprofloxacin (CIP), levofloxacin (LEV), gentamicin (GEN), amikacin (AMK), piperacillin–tazobactam (TZP), fosfomycin (FOS), and imipenem (IMP) (all from Sigma–Aldrich, Shanghai, China). The plates containing the bacterial colonies and the antibiotics were incubated at 37 °C for 18–20 h. The results were interpreted according to CLSI breakpoints published in 2019. Escherichia coli ATCC 25922 cells were used as the quality control strain. The antimicrobial susceptibility of the Shigella isolates was accepted only if the MICs of the quality control strains that were tested in parallel were within the acceptable ranges set by the CLSI guidelines (2019). MDR was defined as resistance to three or more antimicrobial agents.
DNA Extraction and VRG Detection
Bacterial DNA was extracted using the boiling method and plasmid DNA was extracted using the Qiagen Plasmid Purification Kit (QIAGEN, Hilden, Germany). The 16 VRGs (ipaH, ipaA-D, ial, virB, virF, set, sen, icsA, icsB, sigA, sat, pic, and sepA) of the Shigella isolates were identified by polymerase chain reaction (PCR). The primer sequences are shown in Table 1. The cycling conditions included polymerase activation, then an initial denaturation at 95 °C for 5min, followed by 30 cycles of denaturation at 95 °C for 45s, annealing for 45 s (annealing temperatures are shown in Table 1), and extension at 72 °C for 1 min, with a final extension at 72 °C for 10 min. All the PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN) and directly sequenced. Sequences were compared with those of the GenBank nucleotide database to identify the VRGs.
 |
Table 1 Sequences of the Primers Used for PCR Amplification |
Statistical Analysis
Statistical analysis focused on the relationships between Shigella serotype and antimicrobial resistance patterns, and the relationships between VRG profiles and antimicrobial resistance. Data were analyzed using SPSS version 16.0 (SPSS Inc., Chicago, IL, USA). Univariate analysis was performed by the chi-square test or Fisher’s exact test when appropriate. P-values were based on two-tailed test results, and P-values <0.05 were considered statistically significant.
Results
Distribution of Shigella Isolates
A total of 525 non-duplicate Shigella isolates were collected between September 2011 and September 2015. Shigella flexneri was the most abundant (n=449, 85.5%), followed by S. sonnei (n=68, 13.0%). Shigella dysenteriae (n=5) and S. boydii (n=3) were relatively uncommon. Figure 1 shows the yearly distribution of the Shigella isolates. Shigella flexneri was the most frequently isolated species each year. The proportion of S. sonnei isolates increased yearly. Two S. dysenteriae were isolated in 2013 and one in each of 2011, 2012, and 2014. One S. boydii was identified in each of 2012, 2014, and 2015.
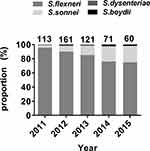 |
Figure 1 The yearly distribution of the proportions of the Shigella isolates. |
Age and Gender
Shigella isolates were collected from specimens from all age groups that presented bacillary dysentery. The median age was 1 year (range: 4 months to 97 years). Infants and children between 1 and 4 years of age accounted for the highest proportion of Shigella isolates (n=331, 63.1%), followed by 5–19-year-old patients (n=72, 13.7%), patients >60 years of age (n=64, 12.2%), and patients between 20 and 59 years of age (n=58, 11.0%) (Figure 2A). Shigella species showed the greatest male predominance. The proportion of S. flexneri was higher in males than in females (57% vs 43%; P<0.0001). There was no significant difference in the proportion of S. sonnei between male and female patients (47.1% vs 53.9%; P=0.49) (Figure 2B).
Antimicrobial Resistance
A comparison of the resistance rates among the Shigella isolates is shown in Table 2. Shigella flexneri was most commonly resistant to AMP (97.3%), followed by NAL (96.9%), TCY (93.8%), and CHL (86.6%). Shigella sonnei was most commonly resistant to AMP (100%), followed by NAL (98.5%), TCY (94.1%), SXT (92.6%), and GEN (92.6%). Shigella flexneri was most sensitive to TZP (99.3%), followed by IPM (98.2%) and AMK (98%). Shigella sonnei was most sensitive to TZP (100%) and AMK (100%), followed by IPM (98.5%) and FEP (98.5%).
 |
Table 2 MIC50 and MIC90 of 17 Antimicrobial Agents Against S. Flexneri and S. Sonnei |
Among the 449 S. flexneri isolates, 417 (92.9%) displayed MDR, and the most common resistance pattern was AMP–NAL–TCY (90.2%). Among the 68 S. sonnei isolates, 66 (94.1%) exhibited MDR, and the most common resistance pattern was AMP–NAL–TCY (92.6%).
Resistance to CHL, CIP, and LVX was more common among S. flexneri isolates than among S. sonnei isolates (86.6% vs 8.8%, P<0.0001; 49.7% vs.19.1%, P<0.0001; 30.5% vs 10.3%, P=0.001, respectively). However, resistance to STX and GEN was more common among S. sonnei isolates than among S. flexneri isolates (76.2% vs 92.6%, P<0.0001; 28.7% vs 92.6%, P<0.0001, respectively).
Prevalence of VRG
Invasion-Associated Genes
The detection of invasion-associated genes among the 449 S. flexneri isolates showed that ipaH had the highest frequency (100%), followed by ipaB (99.8%), ipaC (98.7%), ipaD (98.7%), ipaA (98.4%), and ial (96.0%) (Table 3). The detection of invasion-associated genes among the 68 S. sonnei isolates showed that ipaH had the highest frequency (100%), followed by ipaB (98.5%), ipaC (97.1%), ipaD (95.6%), ipaA (95.6%), and ial (75.0%) (Table 3).
 |
Table 3 Distribution of Virulence-Related Genes Among Shigella Isolates in Anhui, China |
Regulatory Genes
Among the 449 S. flexneri isolates, 439 (97.8%) were positive for the virB gene, while 431 (96.0%) were positive for the virF gene (Table 3). Moreover, a total of 413 isolates (92.0%) were positive for both genes. Among the 68 S. sonnei isolates, 66 (97.1%) were positive for the virB gene, while 51 (75.0%) were positive for the virF gene (Table 3). Fifty (73.5%) isolates were positive for both genes.
Among the 449 S. flexneri isolates, 448 (99.8%) were positive for the icsA gene, while 439 (97.8%) were positive for the icsB gene (Table 3). A total of 437 (97.3%) isolates were positive for both genes. Among the 68 S. sonnei isolates, 67 (98.5%) were positive for the icsA gene and 66 (97.1%) for the icsB gene (Table 3). Moreover, a total of 63 (91.3%) isolates were positive for both genes.
SPATEs
Among the 449 S. flexneri isolates tested, 421 (93.8%) contained class I (sigA, sat) and/or class II (sepA, pic) SPATE genes (Table 3). Moreover, a total of 415 (92.4%) had two class I SPATE genes, and 394 (87.8%) had both class II SPATE genes. The most common SPATE gene among S. flexneri isolates was sat (93.8%), followed by sigA (92.4%), pic (88.2%), and sepA (87.8%). Among the 68 S. sonnei isolates, 64 (94.1%) harbored only one class I SPATE gene (sigA) (Table 3). The sat, pic, and sepA genes were not detected among the S. sonnei isolates in this study.
Enterotoxin Genes
The set gene was present in 420 of the 449 (93.5%) S. flexneri isolates, while the sen gene was present in 413 (92.0%) of the S. flexneri isolates and 60 of the 68 (88.2%) S. sonnei isolates. The set gene was not detected among the S. sonnei isolates in this study.
VRGs and Antimicrobial Resistance
Resistance to CHL and CIP was more common among VRG-positive (ial, ipaA, ipaC, ipaD, virB, virF, set, icsB, sat, pic, and sepA) S. flexneri isolates than among VRG-negative S. flexneri isolates (P<0.05) (Figures 3 and 4). Furthermore, resistance to CRO and CAZ was more common among sat- and set-positive S. flexneri isolates than among sat- and set-negative S. flexneri isolates (P<0.05) (Figure 3E, F). However, CEN resistance was more frequent among VRG-negative (ial, virF, set, sat, pic, and sepA) S. flexneri isolates than among VRG-positive S. flexneri isolates (P<0.05) (Figure 3). With fewer VRGs-negative strains among S. flexneri (ipaH, ipaB, and icsA) and among S. sonnei, no correlation tests were done between these VRGs and antimicrobial resistance. The resistance rates were similar between VRG-positive S. flexneri and VRG-positive S. sonnei isolates, except for the resistance to CRO and CAZ, which was more commonly found among VRG-positive S. flexneri than among VRG-positive S. sonnei (P<0.05) (Supplementary Figure 1 and 2).
Discussion
Shigellosis is an invasive bacterial infection of the human colon that manifests a spectrum of clinical presentations, ranging from short-lasting watery diarrhea to inflammatory bowel disease.3 In this study, we explored the incidence rate of shigellosis during 2011–2015 in Anhui, China, and showed that there was an obvious decline. Shigella flexneri was the most commonly isolated species, similar to that previously reported for several regions in China.16–18 However, we found that the detection rate of S. sonnei, which is the predominant species in developed countries, showed a clear yearly increase. This was likely due to improvements in socioeconomic levels and environmental conditions.18,19
In this study, shigellosis was detected in all age groups; however, children under 5 years of age had higher incidence rates, which was consistent with previous reports.11,17,20 Children under 5 years of age are likely to be more susceptible to shigellosis because of low immune function and the lack of previous exposure.21,22 In this study, we observed that the incidence of S. flexneri was markedly higher among males than among females, similar to that reported in previous studies.16,17 One explanation may be that males have a higher risk of exposure to Shigella-contaminated environments, such as men having sex with men.23 However, there was no significant difference in the incidence of S. sonnei between males and females, which may have been due to the small number of isolated samples.
Effective antimicrobial therapy for shigellosis can significantly relieve symptoms and interrupt bacterial shedding.3 The emergence and dissemination of antimicrobial resistance among Shigella strains in recent years have complicated the therapeutic management of severe shigellosis.6,7,24 Our data showed that Shigella isolates in China had a high resistance to AMP, NAL, TCY, and SXT, suggesting that these drugs can no longer be recommended as empirical therapy for shigellosis. In this study, the Shigella isolates were observed to be susceptible to TZP, IMP, AMK, and FOX, indicating that these antibiotics may be the preferred treatment options in Anhui, China.
In our study, the most frequently detected antimicrobial resistance pattern was AMP–NAL–TCY (90.2% in S. flexneri, 92.6% in S. sonnei), which indicated that MDR among Shigella isolates was widespread in Anhui, China. According to the WHO guidelines for the control of shigellosis, fluoroquinolones and third-generation cephalosporins are effective treatments for this disease.2 However, several studies have reported that the rate of resistance to CIP and CTX in Shigella isolates has gradually increased in recent years,11,25 which was also observed in our study. Of note, our data showed that the susceptibility to CIP and LVX was higher among S. sonnei isolates than among S. flexneri isolates (80.9% vs 50.3%, P<0.0001; 89.7% vs 69.5%, P=0.001, respectively), suggesting that resistance to fluoroquinolones in Anhui, China, was more pronounced for S. flexneri than for S. flexneri. Moreover, the susceptibility to GEN was higher among S. flexneri isolates than among S. sonnei isolates (71.3% vs 7.4%, P<0.0001), suggesting that GEN may be effective as a second-line treatment for S. flexneri infection in Anhui, China.
All Shigella species possess a large virulence plasmid and a single circular chromosome. The T3SS-associated ipa genes are both necessary for the invasion of epithelial cells and the development of shigellosis.26 The ipaH gene, which encodes invasion plasmid antigen H, is commonly used for the molecular identification of Shigella spp. using PCR assays. In our study, all the isolates harbored ipaH, which was consistent with previous reports.27 Most of the Shigella isolates in this study harbored ipaA, ipaB, ipaC, and ipaD, genes that regulate the secretion and translocation of several effector proteins and play a key role in intracellular actin polymerization and depolymerization.28 The ial gene was reported to be responsible for Shigella epithelial cell penetration and cell-to-cell dissemination.29 In our study, the ial gene was present in 96.0% of the S. flexneri isolates and 75.0% of the S. sonnei isolates. Notably, the prevalence of the ial gene in S. sonnei was significantly lower than that in S. flexneri, suggesting that S. sonnei may be less aggressive.
The expression of Shigella virulence genes is regulated by a heat-stable, nucleoid-structuring protein. When the conditions are favorable for invasion, a transcriptional cascade is initiated through the activation of the virF gene, which subsequently turns on the transcription of the regulatory virB gene.30 In this study, 92.0% of the S. flexneri isolates and 73.5% of the S. sonnei isolates were found to harbor both virF and virB, indicating that there might be other pathways for regulating gene expression. Five of the 449 S. flexneri isolates tested possessed virF, but not virB, suggesting that virF may regulate the expression of virulence genes through additional pathways. Interestingly, virB, but not virF, was detected in 3.0% of the S. flexneri isolates and in 21.1% of the S. sonnei isolates, which may have been due to the loss of the virF gene.
IcsB was previously shown to be required for S. flexneri evasion of autophagy at late stages infection (4–6 h) through inhibition of the binding of the autophagy protein Atg5 to the Shigella surface protein IcsA.31 In this study, 97.3% of S. flexneri and 91.3% of S. sonnei harbored both icsA and icsB, indicating that there might be other pathways involved in the prevention of the autophagic recognition of icsA.
The SPATE genes comprise an extended family of secreted autotransporters in Gram-negative bacteria. They exert various functions, including as proteases or mucinases, that can directly or indirectly be toxic to intestine cells.32 In this study, most of the S. flexneri isolates harbored two or more SPATE genes, which is consistent with the results of previous studies.33,34 We also found that the sat gene was the most commonly identified SPATE gene among the S. flexneri isolates, but was absent among the S. sonnei isolates. This indicated that sat-encoded toxin might be a major contributor to the virulence of S. flexneri isolates. Similar to sat, the class II SPATE genes (pic and sepA) were also absent among the S. sonnei isolates in the present study, which indicated that pic and sepA toxins played minor roles in the pathogenicity of S. sonnei.
Two novel enterotoxins (ShET-1 and ShET-2) have recently been described in Shigella isolates. ShET-1 (encoded by set) and ShET-2 (encoded by sen) can alter electrolyte and water transport in the small intestine, which is closely related to the symptoms of dehydration in shigellosis.10,35 In our study, no significant difference was found in the distribution of the sen gene between S. flexneri and S. sonnei. Interestingly, the set gene was not detected among the S. sonnei isolates in this study, suggesting that S. sonnei may be less aggressive than S. flexneri.
This is the first study to demonstrate the correlation between VRGs and antimicrobial resistance among S. flexneri in China. With fewer VRG-negative strains among S. flexneri (ipaH, ipaB, and icsA) and among S. sonnei, no correlation tests were done between these VRGs and antimicrobial resistance. In this study, we observed that resistance to CHL and CIP was more common among VRG-positive (except for sigA and sen) S. flexneri than among VRG-negative S. flexneri; this indicated that, except for sigA and sen, all the S. flexneri VRGs were positively correlated with resistance to CHL and CIP. Furthermore, resistance to CRO and CAZ was more commonly detected among sat- and set-positive S. flexneri isolates than among sat- and set-negative S. flexneri, indicating that the sat and set genes were also positively correlated with resistance to third-generation cephalosporins. Interesting, CEN resistance was more frequent among VRG-negative (ial, virF, set, sat, pic, and sepA) S. flexneri than among VRG-positive S. flexneri, implying that the ial, virF, set, sat, pic, and sepA VRGs in S. flexneri were negatively correlated with CEN resistance.
Conclusion
Our study demonstrated that S. flexneri remains the predominant species in Anhui, China and that the most frequent MDR pattern was AMP–NAL–TCY. This study also highlighted that S. flexneri strains harboring several VRGs were associated with antimicrobial resistance. In summary, we identified the correlation between the VRGs and antimicrobial resistance, thereby improving our understanding of Shigella virulence. Further investigation of VRG expression and the genetic mechanisms underlying the antimicrobial resistance of Shigella isolates is required to help guide empirical antimicrobial therapy for the treatment of shigellosis.
Acknowledgments
This research was supported by research grants from the National Natural Science Foundation of China (No. 81673242 and No. 81973983).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Christopher PR, David KV, John SM, Sankarapandian V. Antibiotic therapy for Shigella dysentery. Cochrane Database Syst Rev. 2010;2010(8):CD006784.
2. Kotloff KL, Winickoff JP, Ivanoff B, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77(8):651–666.
3. Kotloff KL, Riddle MS, Platts-Mills JA, Pavlinac P, Zaidi AKM. Shigellosis. Lancet. 2018;391(10122):801–812. doi:10.1016/S0140-6736(17)33296-8
4. Zhang J, Wang F, Jin H, et al. Laboratory monitoring of bacterial gastroenteric pathogens Salmonella and Shigella in Shanghai, China 2006-2012. Epidemiol Infect. 2015;143(3):478–485.
5. Qiu S, Xu X, Yang C, et al. Shift in serotype distribution of Shigella species in China, 2003-2013. Clin Microbiol Infect. 2015;21(3):252–255.
6. Liu Y, Cheng Y, Yang H, et al. Characterization of Extended-Spectrum beta-Lactamase Genes of Shigella flexneri Isolates With Fosfomycin Resistance From Patients in China. Ann Lab Med. 2017;37(5):415–419.
7. Zhang R, Zhou HW, Cai JC, et al. Serotypes and extended-spectrum beta-lactamase types of clinical isolates of Shigella spp. from the Zhejiang province of China. Diagn Microbiol Infect Dis. 2011;69(1):98–104.
8. Li J, Li B, Ni Y, Sun J. Molecular characterization of the extended-spectrum beta-lactamase (ESBL)-producing Shigella spp. in Shanghai. Eur J Clin Microbiol Infect Dis. 2015;34(3):447–451.
9. Vubil D, Balleste-Delpierre C, Mabunda R, et al. Antibiotic resistance and molecular characterization of shigella isolates recovered from children aged less than 5 years in Manhiça, Southern Mozambique. Int J Antimicrob Agents. 2018;51(6):881–887.
10. Niyogi SK, Vargas M, Vila J. Prevalence of the sat, set and sen genes among diverse serotypes of Shigella flexneri strains isolated from patients with acute diarrhoea. Clin Microbiol Infect. 2004;10(6):574–576. doi:10.1111/j.1469-0691.2004.00897.x
11. Medeiros P, Lima A, Guedes MM, et al. Molecular characterization of virulence and antimicrobial resistance profile of Shigella species isolated from children with moderate to severe diarrhea in northeastern Brazil. Diagn Microbiol Infect Dis. 2018;90(3):198–205. doi:10.1016/j.diagmicrobio.2017.11.002
12. Hazen TH, Leonard SR, Lampel KA, Lacher DW, Maurelli AT, Rasko DA. Investigating the Relatedness of Enteroinvasive Escherichia coli to Other E. coli and Shigella Isolates by Using Comparative Genomics. Infect Immun. 2016;84(8):2362–2371. doi:10.1128/IAI.00350-16
13. Jost BH, Adler B. Site of transcriptional activation of virB on the large plasmid of Shigella flexneri 2a by VirF, a member of the AraC family of transcriptional activators. Microb Pathog. 1993;14(6):481–488. doi:10.1006/mpat.1993.1047
14. Agaisse H. Molecular and Cellular Mechanisms of Shigella flexneri Dissemination. Front Cell Infect Microbiol. 2016;6:29. doi:10.3389/fcimb.2016.00029
15. Parham NJ, Pollard SJ, Desvaux M, et al. Distribution of the serine protease autotransporters of the Enterobacteriaceae among extraintestinal clinical isolates of Escherichia coli. J Clin Microbiol. 2005;43(8):4076–4082. doi:10.1128/JCM.43.8.4076-4082.2005
16. Yang H, Chen G, Zhu Y, et al. Surveillance of antimicrobial susceptibility patterns among Shigella species isolated in China during the 7-year period of 2005-2011. Ann Lab Med. 2013;33(2):111–115. doi:10.3343/alm.2013.33.2.111
17. Chang Z, Zhang J, Ran L, et al. The changing epidemiology of bacillary dysentery and characteristics of antimicrobial resistance of Shigella isolated in China from 2004–2014. BMC Infectious Diseases. 2016;16(1):685. doi:10.1186/s12879-016-1977-1
18. Zhang W, Luo Y, Li J, et al. Wide dissemination of multidrug-resistant Shigella isolates in China. J Antimicrob Chemother. 2011;66(11):2527–2535. doi:10.1093/jac/dkr341
19. Chang Z, Lu S, Chen L, Jin Q, Yang J, Schuch R. Causative species and serotypes of shigellosis in mainland China: systematic review and meta-analysis. PLoS One. 2012;7(12):e52515. doi:10.1371/journal.pone.0052515
20. Sheikh AF, Moosavian M, Abdi M, et al. <p>Prevalence and antimicrobial resistance of Shigella species isolated from diarrheal patients in Ahvaz, southwest Iran. Infect Drug Resist. 2019;12:249–253. doi:10.2147/IDR.S187861
21. Ahmed K, Shakoori FR, Shakoori AR. Aetiology of shigellosis in northern Pakistan. J Health Popul Nutr. 2003;21(1):32–39.
22. Hossain MA, Albert MJ, Hasan KZ. Epidemiology of shigellosis in Teknaf, a coastal area of Bangladesh: a 10-year survey. Epidemiol Infect. 1990;105(1):41–49.
23. Xiao G, Xu C, Wang J, Yang D, Wang L. Spatial–temporal pattern and risk factor analysis of bacillary dysentery in the Beijing–Tianjin–Tangshan urban region of China. Bmc Public Health. 2014;14(1):998. doi:10.1186/1471-2458-14-998
24. Liu Y, Hu L, Pan Y, et al. Prevalence of plasmid-mediated quinolone resistance determinants in association with β-lactamases, 16S rRNA methylase genes and integrons amongst clinical isolates of Shigella flexneri. J Med Microbiol. 2012;61(8):1174–1176. doi:10.1099/jmm.0.042580-0
25. Sadouki Z, Day MR, Doumith M, et al. Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of Shigella sonnei isolated from cases of diarrhoeal disease in England and Wales, 2015. J Antimicrob Chemother. 2017;72(9):2496–2502. doi:10.1093/jac/dkx170
26. Mattock E, Blocker AJ. How Do the Virulence Factors of Shigella Work Together to Cause Disease? Front Cell Infect Microbiol. 2017;7:64. doi:10.3389/fcimb.2017.00064
27. Karimi-Yazdi M, Ghalavand Z, Shabani M, et al. <p>High Rates of Antimicrobial Resistance and Virulence Gene Distribution Among Shigella spp. Isolated from Pediatric Patients in Tehran, Iran. Infect Drug Resist. 2020;13:485–492. doi:10.2147/IDR.S238559
28. Lluque A, Mosquito S, Gomes C, et al. Virulence factors and mechanisms of antimicrobial resistance in Shigella strains from periurban areas of Lima (Peru). Int J Med Microbiol. 2015;305(4–5):480–490. doi:10.1016/j.ijmm.2015.04.005
29. Thong KL, Hoe SL, Puthucheary SD, Yasin R. Detection of virulence genes in Malaysian Shigella species by multiplex PCR assay. BMC Infect Dis. 2005;5:8.
30. Gao X, Zou T, Mu Z, et al. Structural insights into VirB-DNA complexes reveal mechanism of transcriptional activation of virulence genes. Nucleic Acids Res. 2013;41(22):10529–10541. doi:10.1093/nar/gkt748
31. Baxt LA, Goldberg MB, Chakravortty D. Host and bacterial proteins that repress recruitment of LC3 to Shigella early during infection. PLoS One. 2014;9(4):e94653. doi:10.1371/journal.pone.0094653
32. Dautin N. Serine protease autotransporters of enterobacteriaceae (SPATEs): biogenesis and function. Toxins. 2010;2(6):1179–1206. doi:10.3390/toxins2061179
33. Moosavian M, Ghaderiyan GH, Shahin M, Navidifar T. <p>First investigation of the presence of SPATE genes in Shigella species isolated from children with diarrhea infection in Ahvaz, southwest Iran. Infect Drug Resist. 2019;12:795–804. doi:10.2147/IDR.S194740
34. Hosseini NH, Mansouri S, Emaneini M, Moradi M. Distribution of genes encoding virulence factors and molecular analysis of Shigella spp. isolated from patients with diarrhea in Kerman, Iran. Microb Pathog. 2016;92:68–71.
35. Moosavian M, Seyed-Mohammadi S, Sheikh AF, et al. Prevalence of enterotoxin-encoding genes among diverse Shigella strains isolated from patients with diarrhea, southwest Iran. Acta Microbiol Immunol Hung. 2019;66(1):91–101.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.